PhasiRNA is a large class of small RNA in plants, and its function remains to be enveloped. Although the relevant transaction pathways in model plants have been thoroughly studied, further elucidation is required regarding major phasiRNA pathways in non-model plants, including phasiRNA generation from NB-LRR, PPR, MYB and other coding genes, as well as their biological functions. It has been reported that phasiRNA plays an important role in the process of plant reproduction, and their biological functions in the fruit and flower development of horticultural plants warrant further investigation.
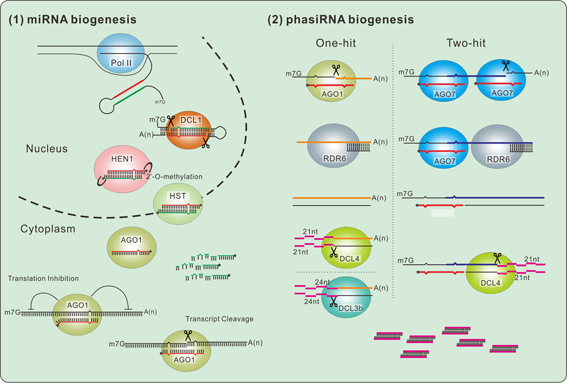
Litchi is one of the most characteristic and dominant large fruit crops in southern China. Taking the advantages of our professional background in small RNA studies and the key research area of fruit science in tropical and subtropical fruit trees of South China Agricultural University, we focus on studying the action pathway of small RNA in litchi.
Firstly, high-throughput deep sequencing technology was used to sequence small RNA from various tissues according to the development stages of litchi.
Subsequently, bioinformatics tools were developed to address the sequencing data analysis problems. Upon the miRNA, miRNA target genes and phasiRNA pathway analysis, an specificly miRNA/phasiRNA metabolic pathways with important biological functions were identified in litchi.
Further studies were conducted on the expression profiles of target miRNA and phasiRNA pathway in various litchi organs at developmental stages or under variable stress conditions.
Secondly, similar techniques were used to conduct exploratory studies on small RNA in other representative tropical and subtropical fruit trees.
By comparing across species, the conserved and unique regulation pathways of small RNA metabolism in tropical and subtropical fruit trees were identified. Finally, functional verification experiment was carried out in the strawberry model system.
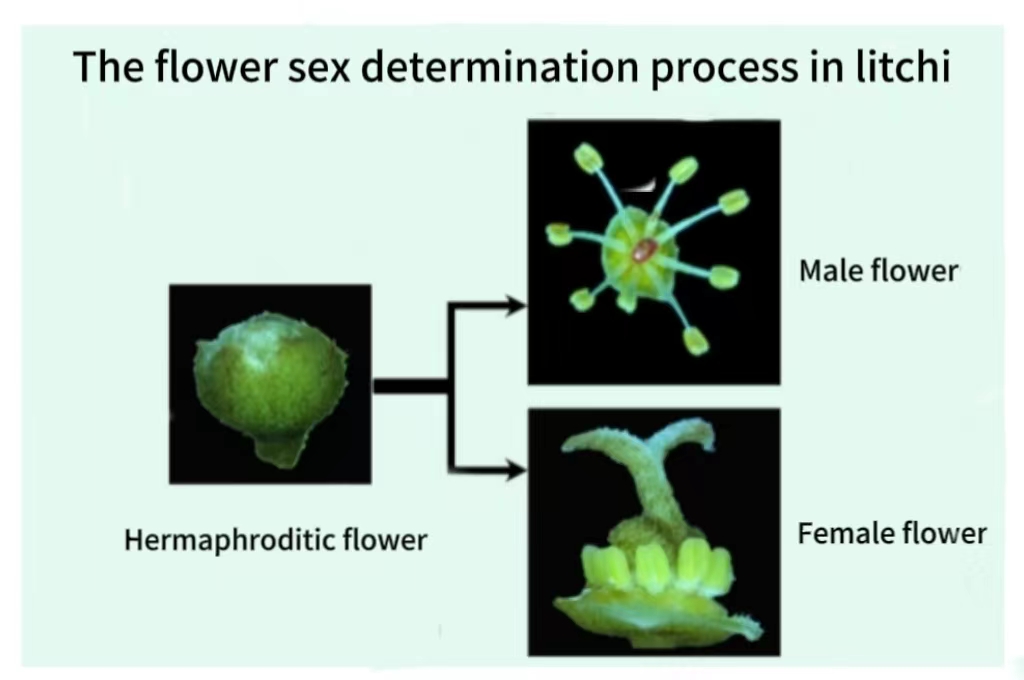
Flower development and sexual determination represent the most valuable and challenging research areas in relation to crop production for fruit harvesting purposes. Current research suggests that small RNA also play a critical role in this important pathway. For example, miR172 regulates flower development by acting on AP2 transcription factor, while the auxin response factor that miR390, miR160 and miR167 work together plays an important role in flower development and fertility. Recent research on persimmon, a woody fruit tree, has revealed that a small RNA located on the Y chromosome plays a crucial role in determining flower sex. Population genetics analysis in rice has revealed that a phototherm-sensitive male sterile gene is a PHAS site (phasiRNA production) acting on miR482/2118.
Tropical and subtropical fruit trees exhibit diverse developmental forms of flowers, with varying sexual characteristics. For instance, male, female, and hermaphroditic flowers are present in litchi and longan, each exhibiting distinct flowering periods. Papaya plants show similar floral sexes including male, female and hermaphrodite. We employ a combination of genetic, comparative genomics, cell biology, bioinformatics and various high-throughput deep sequencing techniques to investigate the mechanisms underlying flower development and sexual determination in representative tropical and subtropical fruit trees such as litchi, longan and papaya. Our research focuses on elucidating the role of small RNA in these processes, providing a theoretical foundation for effective regulation of flower development and fruit formation in agricultural production.
The unique tropical and subtropical climate in South China has fostered a diverse range of horticultural crops. Using high-throughput sequencing technology and independent analysis of biological information data, our research group aims to sequence the genomes of major horticultural crops with southern characteristics and establish a genome data platform. In addition, extensive collection and utilization of abundant germplasm resources and a series of means such as resequencing technology were used to identify the key regulatory factors for the formation of major economic traits of these crops. Currently, our research group has participated in genome sequencing of litchi, longan and other major fruit crops.
With the advent and rapid advancement of high-throughput sequencing technology, biological research has entered the era of "big data". The effective analysis of these large-scale biological datasets relies on continuous optimization of analytical tools and convenient sharing of data. Based on the existing foundation, our research group will focus on developing and optimizing various data analysis tools, particularly those related to small molecule RNA sequencing data. Our goal is to achieve a more user-friendly interface for data analysis and visualization.
Copyright © 2023.XIALAB, College of Horticulture, South China Agricultural University Address : Room 913, College of Horticulture, South China Agricultural University, Wushan,No. 483 Wushan Road,Tianhe District, Guangzhou, Guangdong Province